Our researched is focused on biological soft matter: its dynamics, conformational transitions, and processes leading to the hierarchical self-assembly of biopolymers and biomembranes. We are particularly interested in:
- mechanisms of protein aggregation,
- borderlines between determinism and randomness in conformational transitions,
- thermodynamics of protein folding with particular emphasis on high pressure effects,
- applications of the biological self-organization in nanotechnology.
A single unifying aspect of the different research topics studied in our group is the physicochemical common-sense-approach applied to the self-assembly of life on its molecular and microscopic levels. Paradoxically, misfolded and aggregated proteins – which in vivo are associated with disease and demise, rather than blooming life – have turned out to be very revealing models in this regard. Aggregation of protein molecules and formation of a certain type of proteinaceous nanofibrils, the so-called amyloids have been linked to a number of fatal disorders such as Alzheimer, Parkinson, or Creutzfeldt-Jakob Diseases (the prion-related illness). Presently, molecular mechanisms responsible for transition of correctly folded biologically-functional protein molecules into their toxic assemblies remain poorly understood. This is the main obstacle that prevents finding successful cures for such maladies. Yet, as the phenomenon of protein aggregation appears to reflect a common generic feature of proteins as polymers, it also draws a lot of analogies to other physical processes, which at first glance appear entirely ‘unbiological’.
Finding such analogies between the biology that needs mending and phenomena from the realm of condensed matter physics stimulates our studies of structural and thermodynamic mysteries of protein aggregation. We focus on model proteins (e.g insulin, alpha-lactalbumin, lysozyme), sequenceless polypeptides (polylysine and polyglutamic acid), using the whole plethora of biophysical methods including FT-IR/Raman spectroscopy, fluorescence, circular dichroism, AFM/SEM/TEM microscopic techniques and many others.
The specific areas of our interests are outlined along following ‘big’ questions that remain to be answered:
- What is the exact sequence of events on misfolding/aggregation pathways of proteins? Can we come up with a way of preventing these processes in vivo using genomic and non-genomic means?
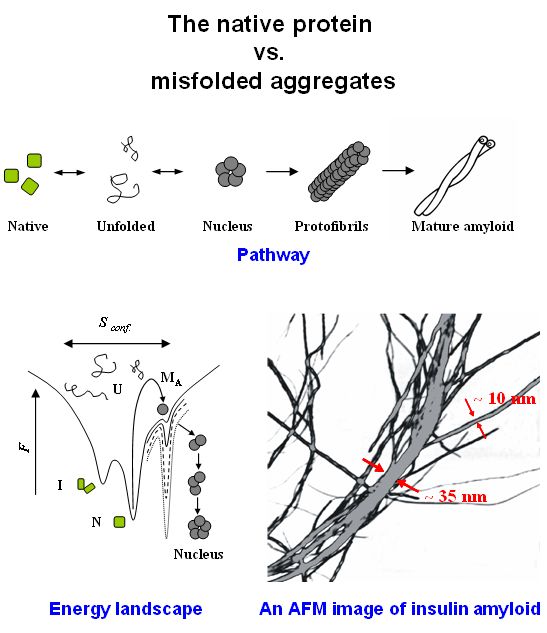
A simplified representation of a protein aggregation pathway.
Jansen R, Dzwolak W, Winter R “Amyloidogenic self-assembly of insulin aggregates probed by high resolution atomic force microscopy” Biophys. J. 88 (2005) 1344-1353.
- A related question: What are the molecular mechanisms underlying the memory effect observed in trans-generational (mother → daughter) proliferation of conformational variants of amyloid fibrils? This problem appears to be closely related to the so-called ‘prion strains phenomenon’, and as-yet obscure conformational origins of toxicity of misfolded proteins.
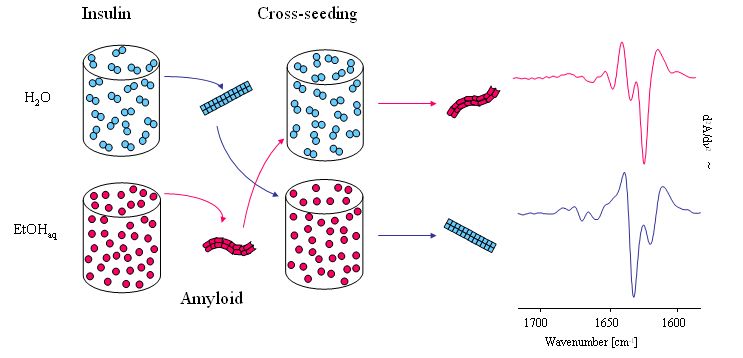
The ‘memory effect’ reflects the strain-dependent amyloidogenesis of insulin
Dzwolak W, Smirnovas V, Jansen R, Winter R “Insulin Forms Amyloid in a strain-dependent manner: an FT-IR spectroscopic study” Protein Sci. 13 (2004) 1927-1932,
Dzwolak W, Jansen R, Smirnovas V, Loksztejn A, Porowski S, Winter R “Template-controlled conformational patterns of insulin fibrillar self-assembly reflect history of solvation of the amyloid nuclei” Phys.Chem.Chem.Phys. 7 (2005) 1349-1351.
Dzwolak W, Grudzielanek S, Smirnovas V, Ravindra R, Nicolini C, Jansen R, Loksztejn A, Porowski S, Winter R “Ethanol-perturbed amyloidogenic self-assembly of insulin: looking for origins of amyloid strains” Biochemistry 44 (2005) 8948-8958.
- What role does chaos play in protein misfolding? Are hydrodynamic forces responsible for symmetry-breaking in aggregating insulin? These questions follow the surprising discovery of the chiral bifurcation phenomenon, which accompanies – under certain conditions – aggregation of insulin.
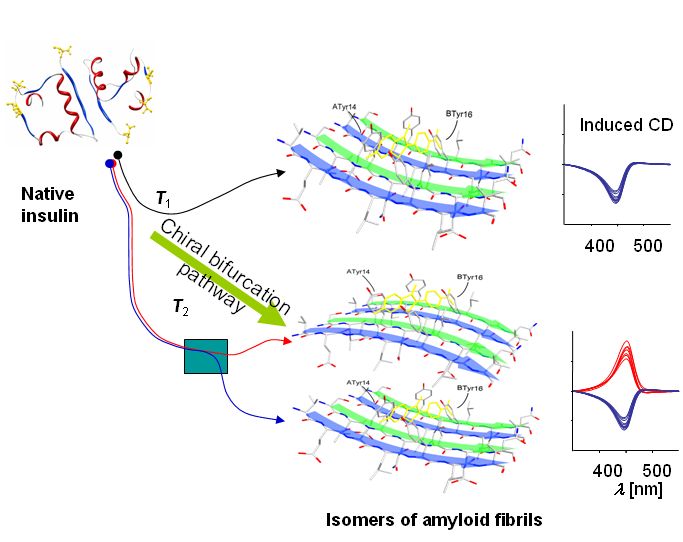
Chiral bifurcation -
upon agitation, insulin stochastically chooses to convert to either of two chiral variants of amyloid. Microscopic fluctuations determine symmetry of a macroscopic amount of aggregate.
Loksztejn, A, Dzwolak, W, “Chiral bifurcation in aggregating insulin: An induced circular dichroism study” J. Mol. Biol., 379 (2008) 9-16.
Dzwolak W, Loksztejn A, Galinska-Rakoczy A, Adachi R, Goto Y, Rupnicki L, “Conformational indeterminism in protein misfolding: chiral amplification on amyloidogenic pathway of insulin” J. Amer. Chem. Soc. 129 (2007) 7517-7522.
Dzwolak W, Pecul M “Chiral bias of amyloid fibrils revealed by the twisted conformation of thioflavin T: an induced circular dichroism / DFT study” FEBS Lett. 579 (2005) 6601-6603.
- How conditions, that are extremely hostile to the native state, may affect self-assembly of proteins, and what this can teach us about the nature of forces driving folding and misfolding?
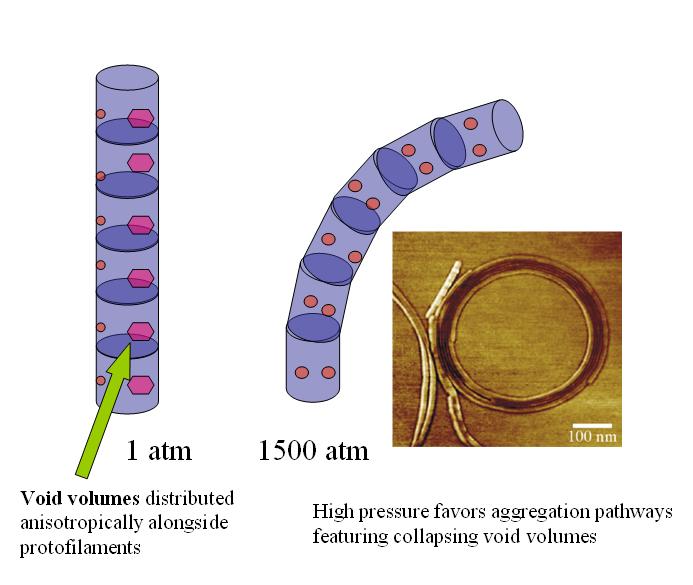
Unique circular insulin fibrils are formed under high pressure, suggesting an anisotropic distribution of void volumes within the amyloid.
Jansen R, Grudzielanek S, Dzwolak W, Winter R, “High Pressure Promotes Circularly Shaped Insulin Amyloid” J. Mol. Biol., 338 (2004) 203-206.
- What is the nature of docking interactions between amyloid fibrils and small organic and inorganic ligands?
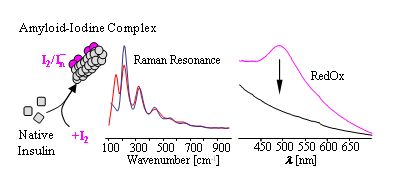
Amyloids form inclusion complexes with molecular iodine, much the same way starch does.
Dzwolak W “Insulin amyloid fibrils form an inclusion complex with molecular iodine: a misfolded protein as a nanoscale scaffold” Biochemistry 46 (2007) 1568-1572.
- Another take from the bionanotechnological perspective: Could useful ‘molecular devices’ be assembled from protein fibrils as building blocks?
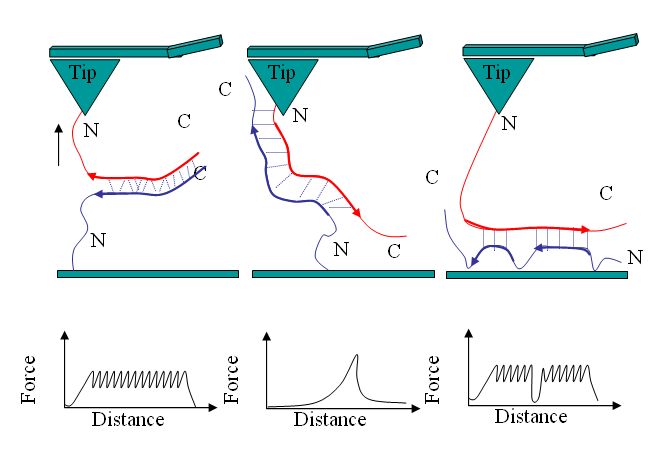
‘Molecular Velcro’–like properties of a beta-pleated heterostructure formed by poly-L-lysine and poly-L-glutamic acid.
Dzwolak W, Marszalek PE, “Zipper-like properties of [poly(L-lysine)+poly(L-glutamicacid)] beta-pleated molecular self-assembly” Chem. Commun. 44 (2005) 5557-5559.
Dzwolak W “Insulin amyloid fibrils form an inclusion complex with molecular iodine: a misfolded protein as a nanoscale scaffold” Biochemistry 46 (2007) 1568-1572.